paired end sequencing insert size
Paired-end RNA sequencing RNA-Seq enables discovery applications such as detecting gene fusions in cancer and characterizing novel splice isoforms. Detection and characterization of novel sequence insertions using paired-end next-generation sequencing.

Pdf Assessment Of Insert Sizes And Adapter Content In Fastq Data From Nexteraxt Libraries
For instance for a PE150 sequencing run the insert size should be above 300bp.

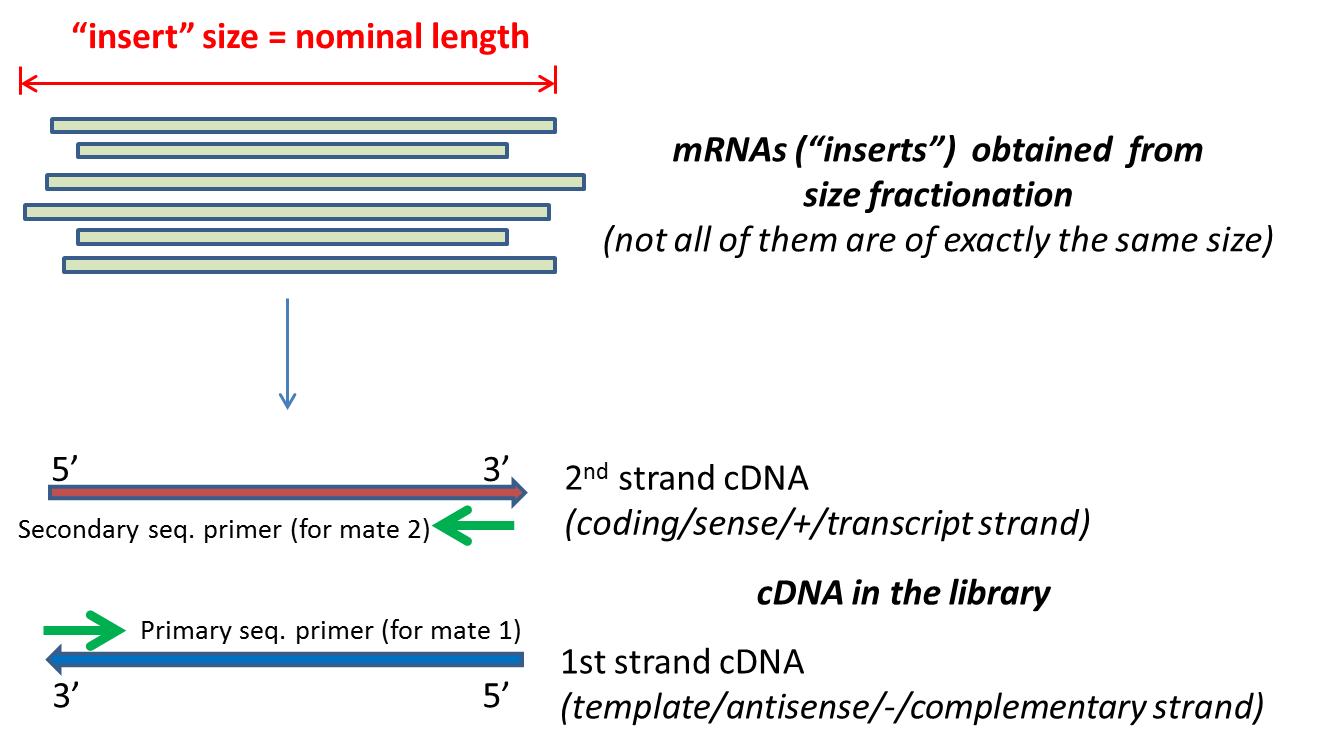
. The insert is normally the stretch of sequence between the paired-end adapters so in your case the insert size would be 250 bp 2x75 bp reads 100 bp unsequenced middle piece. Smaller DNA fragments are aligned to the largest DNA fragments by one of their two-ends. By paired-end sequencing of a series of stepwise insert size libraries we are able to recover the full length sequences of the largest DNA fragments using computational method.
2Align paired-end RNA-Seq data to the reference transcripts using minimum insert-length as zero and maximum insert-length as 500 or more. Such reads will also contain adapter sequences which may need to be trimmed they as can negatively impact on some types of. PE reads generally have a smaller insert size 1kp than MP 2-5 kb.
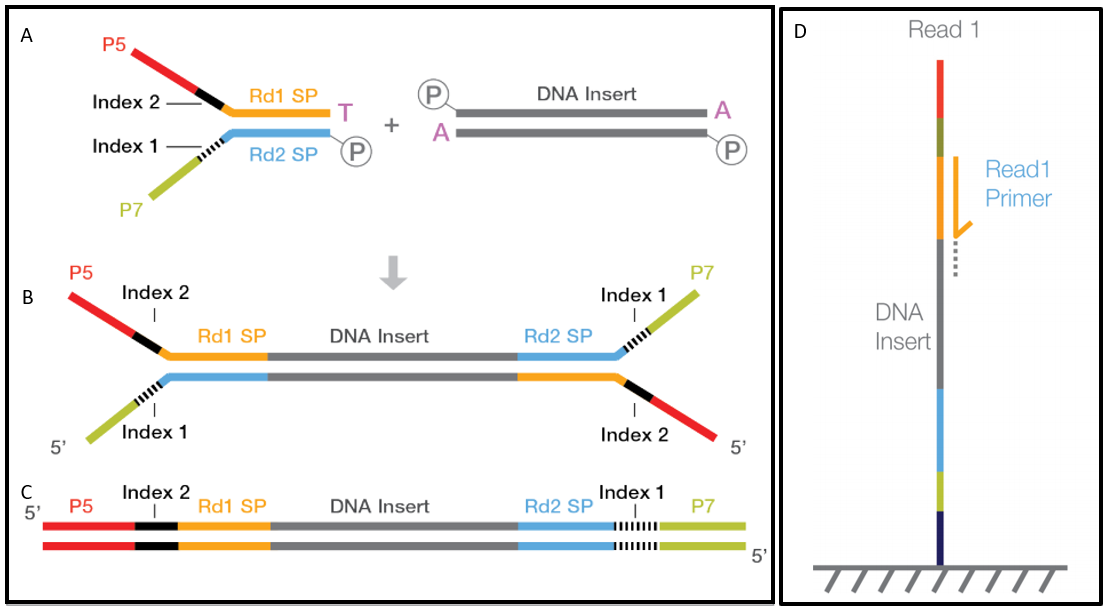
Depending on the length of your reads it is possible for PE reads to have overlapping ends. PE reads generally have a smaller insert size 1kp than MP 2-5 kb. The sequencing starts at Read 1 Adapter mate 1 and ends with the sequencing from Read 2 Adapter mate 2.
Thats why you should first take a look in the operating instructions of your used device and kit. However the detectable sequence length with read-pair analysis is limited by the insert size. For PE reads insert size refers to the fragment size excluding the adapters.
However the average size of. Paired end sequencing refers to the fact that the fragment s sequenced were sequenced from both ends and not just the one as was true for first. -I 0 -X 500 BWA.
Thus the other ends can be used to fill the un-sequenced regions in the largest DNA fragments. A Sample-wise IS histograms of liquid biopsies LBs. Im no expert in library preparation for DNA sequencing but as I understand it protocols for paired-end and mate-pair sequencing involve a size selection step that selects DNA fragments within a tight target size range.
For this reason there is no general recommendation and we can only give. The reads have a length of typically 50 - 300 bp. Ranges from 2 150 to 2 300 bp.
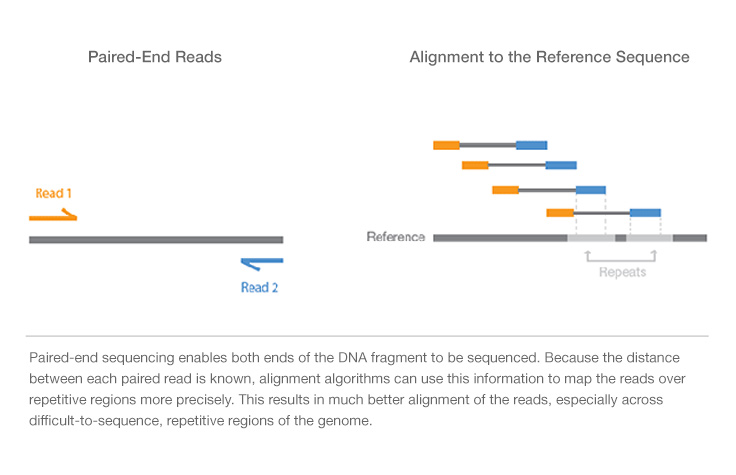
A computational framework to discover. The algorithm is simple. Paired end reads with an insert size of less than the length of a single read contain less information than read pairs with longer insert sizes.
Department of Energys Office of Scientific and Technical Information. Including flanking intronic regions of. Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired.
The inner mate distance between the two reads is 200 bp creating an insert size of 400 bp. You can use sequencing reagents to generate single continuous reads or for. B Histology-wise mean IS.
Department of Energys Office of Scientific and Technical Information. In an Illumina sequencing run either single-end sequencing SE or paired-end sequencing PE can be used. For mRNA-Seq library prep use.
In any case DNA is chopped into small fragments ligated with adapters at both ends and sequenced either from one end SE or from both ends PE. Thus longer sequence insertions that contribute to our genetic makeup are not extensively researched. Normally the insert size is longer than the sum of the two read lengths meaning there is an unsequenced inner part in the middle of the insert.
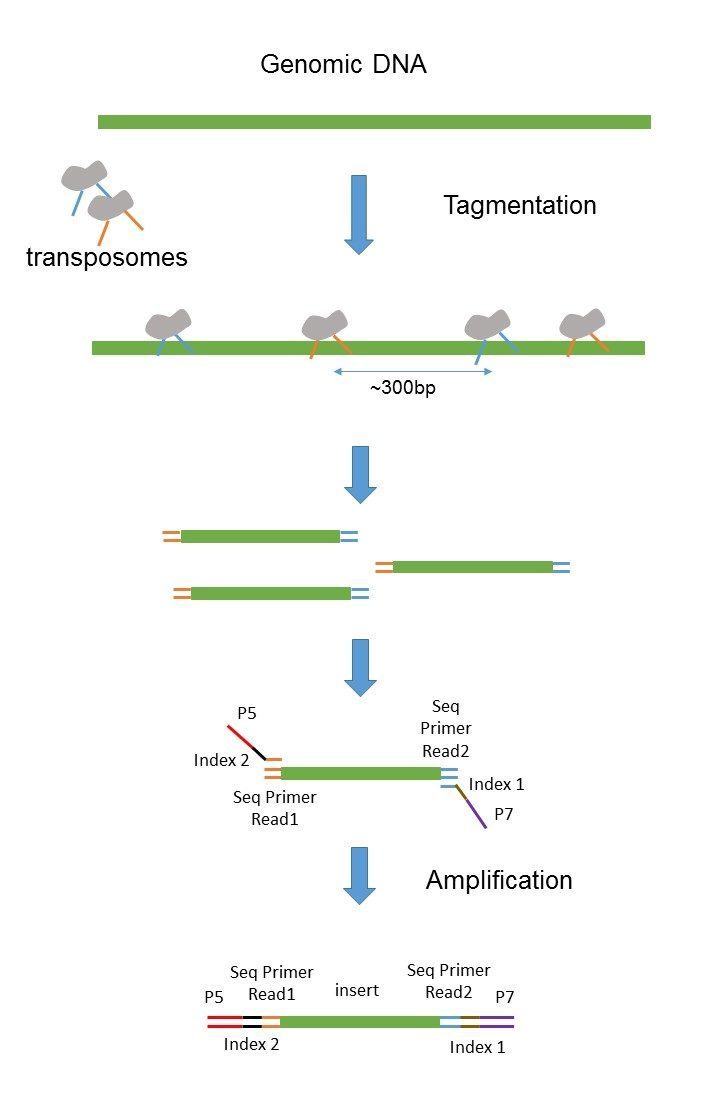
Insert size -- The insert size refers to the distance between the pairs. It depends on the used Illumina system the used reagents kits and the mode singlepaired end. By controlling the size of sequenced DNA fragments or inserts borrowing terminology from the days when DNA fragments were actually.
Check the TLEN field in the SAM format throw out pair-end reads whose pairs are too far away and use them to estimate the mean and variance of the insert length. You can use sequencing reagents to generate single continuous reads or for paired-end sequencing in both directions. The unique paired-end sequencing protocol allows the end user to choose the length of the insert 200500 bp and sequence either end of the insert generating highly quality alignable sequence data.
By computational modeling we show how libraries of multiple insert sizes combined with strand-specific paired-end SS-PE sequencing can increase the information gained on alternative splicing especially in higher eukaryotes. For paired-end RNA-Seq use the following kits with an alternate fragmentation protocol followed by standard Illumina paired-end cluster generation and sequencing. For example a 300-cycle kit can be used for a 1 300 bp single-read run or a 2 150 bp paired-end run Beginners Guide to Next-Generation Sequencing.
For paired-end RNA-Seq use the following kits with an alternate fragmentation protocol followed by standard Illumina paired-end cluster generation and sequencing. Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a single insert size. The name insert as.
I wrote a single Python script to estimate the paired-end read insert length or fragment length from read mapping information ie SAMBAM files. The reads were then mapped back to the reference using BWA aln and sampe. The difference in insert size stems from the difference in protocols.
However the average size of human coding exons is only 160bp. If they occur at significant levels the amount of useable sequence in a dataset will be reduced. What is the optimal insert size.
1Create reference transcriptome index for BowTie or BWA. For Illumina systems DNA insert size range of 200800 bp. One could argue that longer read length generates more output from the same amount of input molecules therefore the insert size should be increased accordingly to make use of this extra data.
Download scientific diagram Paired-end PE sequencing and insert size IS filtering to increase sensitivity.

What Are Paired End Reads The Sequencing Center

What Is Mate Pair Sequencing For
How Short Inserts Affect Sequencing Performance
What Is Paired End 150 Omega Bioservices

Interpreting Color By Insert Size Integrative Genomics Viewer
The Insert Size In Paired End Data Seqanswers
Paired End Sequencing France Genomique

What Is Mate Pair Sequencing For

Definition Of Essential Terms A Definition Of Fragment Read And Download Scientific Diagram

What Are Paired End Reads The Sequencing Center

Diagram To Show The Construction Of A Fragment With An Insert Size Is Download Scientific Diagram
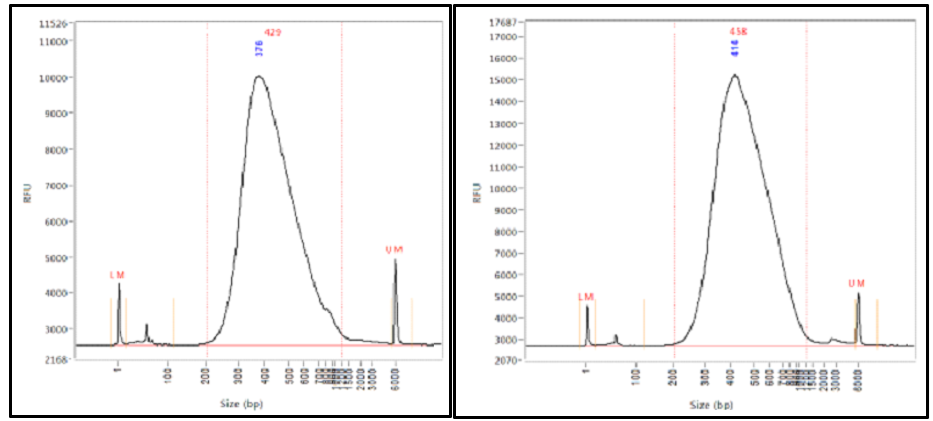
How Short Inserts Affect Sequencing Performance
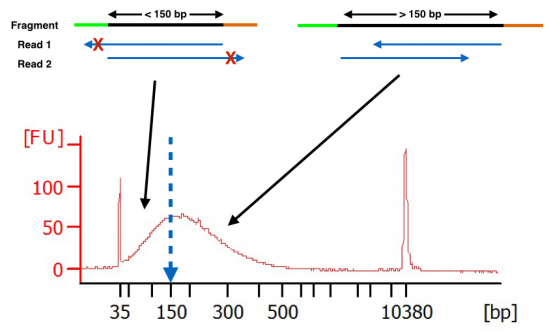
What Happens When Hardware Advances Faster Than Molecular Protocols Cofactor Genomics

Diagram To Show The Construction Of A Fragment With An Insert Size Is Download Scientific Diagram

Interpreting Color By Insert Size Integrative Genomics Viewer
What Read Lengths Are Produced By Modern Illumina Sequencers